De, S., Stadlmayr, G., Rebnegger, C., Mattanovich, D., & Gasser, B.
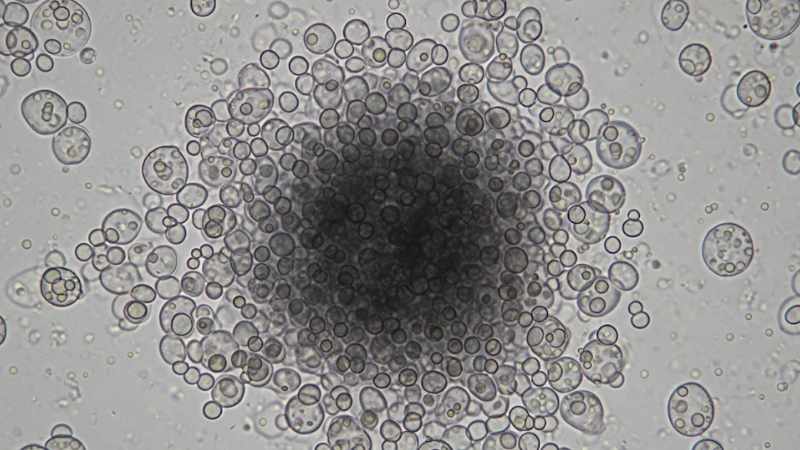
The yeast Komagataella phaffii (Pichia pastoris) is crucial in modern biotechnology, especially for producing proteins. Controlling yeast cell behavior is vital for efficient industrial processes. One key behavior is flocculation – where cells clump together and settle. When controlled, this can significantly simplify product separation, reducing costs. Researchers from leading Austrian institutions, including the Austrian Centre of Industrial Biotechnology (acib) and BOKU University Vienna, have made a significant discovery about K. phaffii‘s flocculation. Published in Applied Microbiology and Biotechnology, their study uncovers a fascinating pH-dependent clumping mechanism with major industrial implications.
A pH-controlled clumping switch
K. phaffii cells clump most effectively at a mildly acidic pH of 4.0. This clumping is reversible: changing the pH disperses flocs. Calcium ions accelerated clumping, indicating flocculin involvement. This precise pH-responsive clumping offers a new, simple control mechanism.
Surprising gene roles and unique mechanisms
The study investigated the genetic factors behind this phenomenon. Six flocculin (FLO) genes showed altered activity at low pH. The gene Flo5-1 was particularly surprising: its deletion actually increased flocculation, suggesting it normally inhibits clumping or acts as a sensing protein.
Crucially, the study identified Nrg1, a transcriptional repressor, as a key player. Nrg1 was found to negatively regulate pH-dependent flocculation; overexpressing Nrg1 significantly reduced clumping. Nrg1 acts as a powerful genetic switch. Unlike Saccharomyces cerevisiae, K. phaffii‘s Flo8, a key regulator in other yeasts, had no role in this process. This highlights the need to study non-conventional yeasts directly, as their biological pathways can differ.